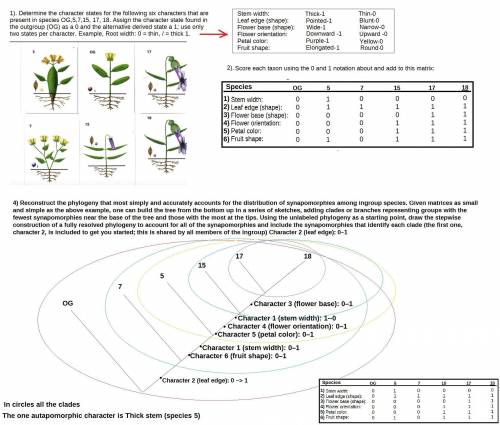
Reconstruct the phylogeny that most simply and accurately accounts for the distribution of synapomorphies among ingroup species. Given matrices as small and simple as the above example, one can "build" the tree from the bottom up in a series of sketches, adding clades or branches representing groups with the fewest synapomorphies near the base of the tree and those with the most at the tips. Using the unlabeled phylogeny as a starting point, draw the stepwise construction of a fully resolved phylogeny to account for all of the synapomorphies, and include the synapomorphies that identify each clade (the first one, character 2, is included to get you started; this is shared by all members of the ingroup).

Answers: 3


Another question on Biology

Biology, 21.06.2019 15:00
Although there are a limited number of amino acids many different types of proteins exist because..
Answers: 1

Biology, 22.06.2019 07:00
Give two examples of what would occur if one of the pairs did not match or had an extra chromosomes. explain how this might occur, and if it would be dangerous to the individual. fast.
Answers: 1

Biology, 22.06.2019 08:30
Gene expression is the activation of a gent that results in a question 1 options: protein dna mitochondria
Answers: 1

Biology, 22.06.2019 10:00
Suppose you use three different scale to weigh a bag of oranges. one scale says the nag weighs 2.1 lb, and third says it weighs 2.1 lb. the actual weight of the bag of oranges is 2.153 lb. which of the following best decribes these results?
Answers: 2
You know the right answer?
Reconstruct the phylogeny that most simply and accurately accounts for the distribution of synapomor...
Questions

History, 28.07.2019 22:40

Biology, 28.07.2019 22:40

Business, 28.07.2019 22:40

Mathematics, 28.07.2019 22:40

Mathematics, 28.07.2019 22:40

Mathematics, 28.07.2019 22:40

Mathematics, 28.07.2019 22:40

Physics, 28.07.2019 22:40




Mathematics, 28.07.2019 22:40

History, 28.07.2019 22:40

Mathematics, 28.07.2019 22:40



Chemistry, 28.07.2019 22:40

History, 28.07.2019 22:40

Social Studies, 28.07.2019 22:40

History, 28.07.2019 22:40